What Kevin McKernan has been warning about has been confirmed
N1-methylpseudouridine (m1Ψ) into mRNA results in +1 ribosomal frameshifting
Let’s first define what ribosomal frameshifting is:
(From Wiki) Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA.
This process is naturally regulated and can increase the coding possibilities for cells and viruses in ways that are not well understood. From (Ref 1):
Frameshifting of mRNA during translation provides a strategy to expand the coding repertoire of cells and viruses. How and where in the elongation cycle +1-frameshifting occurs remains poorly understood.
With regard to the introduction of N1-methylpseudouridine into mRNA it was already clear within the literature that similar transformations, such as mRNA pseudouridylation (Ψ), are naturally regulated in a very coordinated manner in order to enhance the possibilities of the genetic code, modulating and even re-writing its expression. From (Ref 2):
Notably, the majority of pseudouridines in mRNA are regulated in response to environmental signals, such as nutrient deprivation in yeast and serum starvation in human cells. These results suggest a mechanism for the rapid and regulated rewiring of the genetic code through inducible mRNA modifications.
Because Ψ stabilizes RNA structure, mRNA pseudouridylation could alter translation initiation efficiency, ribosome pausing, RNA localization, and regulation by RNA interference, to name a few aspects of mRNA metabolism known to be affected by RNA structure, although we cannot exclude the possibility that many instances of mRNA pseudouridylation may be functionally silent. However, given recent evidence that pseudouridine profoundly affects decoding by ribosomes from diverse organisms, our results also raise the possibility of widespread regulated rewiring of the genetic code.
Therefore there are no surprises coming from the recent publication titled [N1-methylpseudouridylation of mRNA causes +1 ribosomal frameshifting] (Ref 3). As the authors themselves indicate, we are working within an uncharted territory:
Modified ribonucleotides are commonly incorporated into therapeutic IVT mRNAs to decrease their innate immunogenicity, but their effects on mRNA translation fidelity have not been fully explored.
Similar findings have already been reported in the case of Ψ and m1Ψ, as Kevin McKernan pointed out in a recent interview with the Epoch times (Ref 4). In one study (Ref 5), unusual based pairing was found in the decoding of the stop codon, interrupting the termination of protein synthesis:
During normal translation, binding of a release factor to one of the three stop codons (UGA, UAA or UAG) results in termination of protein synthesis. However, modification of the initial uridine to a pseudouridine (Ψ) allows efficient recognition and read-through of these stop codons by a transfer RNA (tRNA), although it requires formation of two normally forbidden purine-purine base pairs.
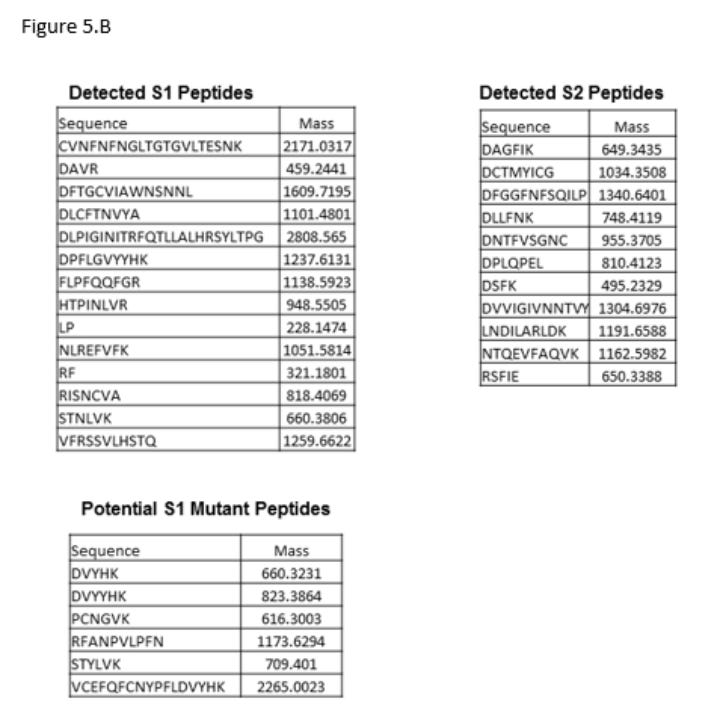
In another study (Ref 6), also pointed out by Kevin McKernan, patients following SARS-CoV-2 vaccines presented mutant S1 peptides, which could be a direct manifestation of translational errors during the transfection process. From the study:
Further analysis revealed that these S1 positive, CD16+ cells also contained peptide sequences of S2, and mutant S1 peptides (Fig 5B).
For more clarification on this phenomena we can watch a segment of Kevin McKernan’s interview at The Epoch Times (Follow link to Ref 4 for the full interview):
My Conclusion:
We already knew about the potential, and I quote, “translation fidelity issues” with this modRNA products, as Kevin McKernan has pointed out several times in different platforms. It also was already very clear within the scientific literature. For this reason alone, experiments such as the one presented in (Ref 3) (i.e. the study upon which this whole article is based) should have been required before deploying these highly experimental products in the population.
This proves, once again, that modRNA is an unstable drug that cannot be properly tested for safety and efficacy in its current formulation, as it would create random distributions of peptides and potentially cause severe side effects in otherwise healthy individuals. This constitutes the opposite of a therapeutic, where a reliable knowledge on dosage, distribution, duration, reproducibility, fidelity, stability, integrity of the components, purity... is needed. None of this have been achieved so far in the application of this technology for immunization purposes.
The biodistribution studies for the LPNs are another example of this same phenomena. From Acuitas Therapeutics Inc. (Ref 7), which is the provider of LPNs for the mRNA vaccines that have been deployed:
In conclusion, the distribution of [3H]-08-A01-C01 (monitoring the [3H]-CHE lipid label) in blood, plasma and selected tissues was determined in male and female Wistar Han rats over 48 hours after a single intramuscular injection at 50 µg mRNA/animal (1.29 mg/animal lipid dose). The concentrations of [3H]-08-A01-C01 were greatest in the injection site at all time points, with levels peaking in the plasma by 1-4 hours post-dose and distribution mainly into liver, adrenal glands, spleen and ovaries over 48 hours. Total recovery of radioactivity outside of the injection site was greatest in the liver, with much lower total recovery in spleen, and very little recovery in adrenals glands and ovaries. The mean plasma, blood and tissue concentrations and tissue distribution patterns were broadly similar between the sexes and [3H]-08-A01-C01 did not associate with red blood cells.
From the same report (Ref 7),
The highest mean concentrations observed, and the equivalent % dose, are presented in the tables below.
Therefore we just cannot differentiate between a toxic and a therapeutic dosage if transfection efficiency and biodistribution, as well as toxicity and concentration levels of by-products, are unknown factors hiding behind random distributions that behave differently in different cohorts of the population as well as from individual to individual depending on unpredictable metabolic and environmental factors. That is, even in the absence of contaminants (such as fragments of plasmid DNA coming from the manufacturing process), we still do not have a reliable, reproducible effect that can be properly tested as a therapeutic; especially in a cohort of otherwise, healthy individuals.
Aside note: Such considerations may not be equally relevant in the field of cancer treatment, or genetic disorders, due to the considerably different risk/benefit ratio.
REFERENCES:
Ref 1: Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation https://www.nature.com/articles/s41467-021-24911-1
Ref 2: Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells https://www.nature.com/articles/nature13802
Ref 3: N1-methylpseudouridylation of mRNA causes +1 ribosomal frameshifting https://www.nature.com/articles/s41586-023-06800-3?utm_source=substack&utm_medium=email
Ref 4: Kevin McKernan Talks COVID Vaccine DNA Contamination, the Monkey Virus SV40 Promoter, and What’s Actually in the Vaccines https://www.theepochtimes.com/epochtv/kevin-mckernan-talks-covid-vaccine-dna-contamination-the-monkey-virus-sv40-promoter-and-whats-actually-in-the-vaccines-5481974
Ref 5: Unusual base pairing during the decoding of a stop codon by the ribosome https://www.nature.com/articles/nature12302
Ref 6: SARS-CoV-2 S1 Protein Persistence in SARS-CoV-2 Negative Post-Vaccination Individuals with Long COVID/ PASC-Like Symptoms https://www.researchsquare.com/article/rs-1844677/v1
Ref 7: A Tissue Distribution Study of a [3 H]-Labelled Lipid Nanoparticle-mRNA Formulation Containing ALC-0315 and ALC-0159 Following Intramuscular Administration in Wistar Han Rats https://icandecide.org/wp-content/uploads/2022/03/125742_S1_M4_4223_185350.pdf