Following my previous substack [The real problem with Whole Genome Sequencing (WGS)] I have changed the methodology of my experiment in order to caclulate the error rate of WGS for the SARS-CoV-2 phylogenetic trees. It seems that it will take a while, there are many techniques and software that I have to learn about in order to do this with sufficient precision.
SOME REFERENCES THAT I THINK ARE IMPORTANT:
"Tiled amplicon RT-PCR followed by sequencing with Illumina or Oxford Nanopore technology is the most popular method for generating SARS-CoV-2 whole genome sequences." (Ref. 1). For PCR tiling go to (Ref. 2)
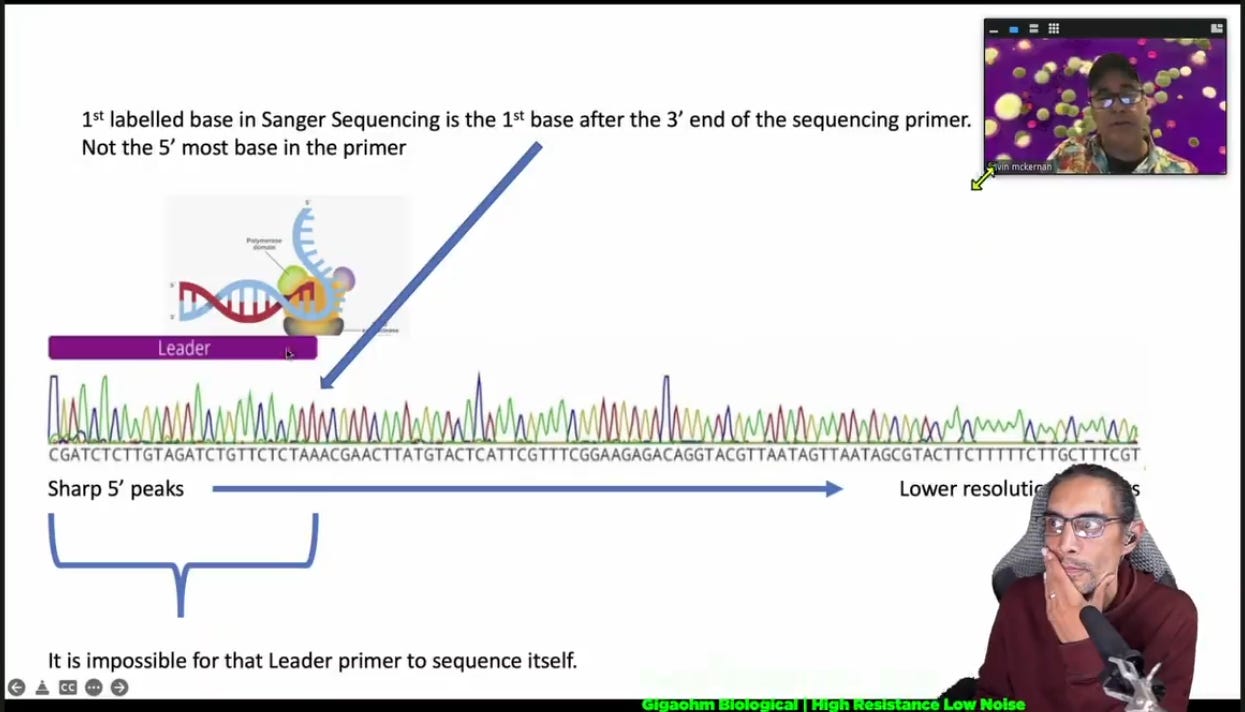
Would sequencing of tiled PCR-generated amplicon pools create an accurate phylogenetic tree? Can it capture recombination events? I don't think so... As I said I am trying to calculate the error rate. It will take a while... For additional reference on the subject I would encourage the reader to watch this video from Gigaohm Biological, (Kevin McKernan LIVE: Gigaohm Biological High Resistance Low Noise Information Brief). The video has also been store (Here), in case the previous link fails in the future.
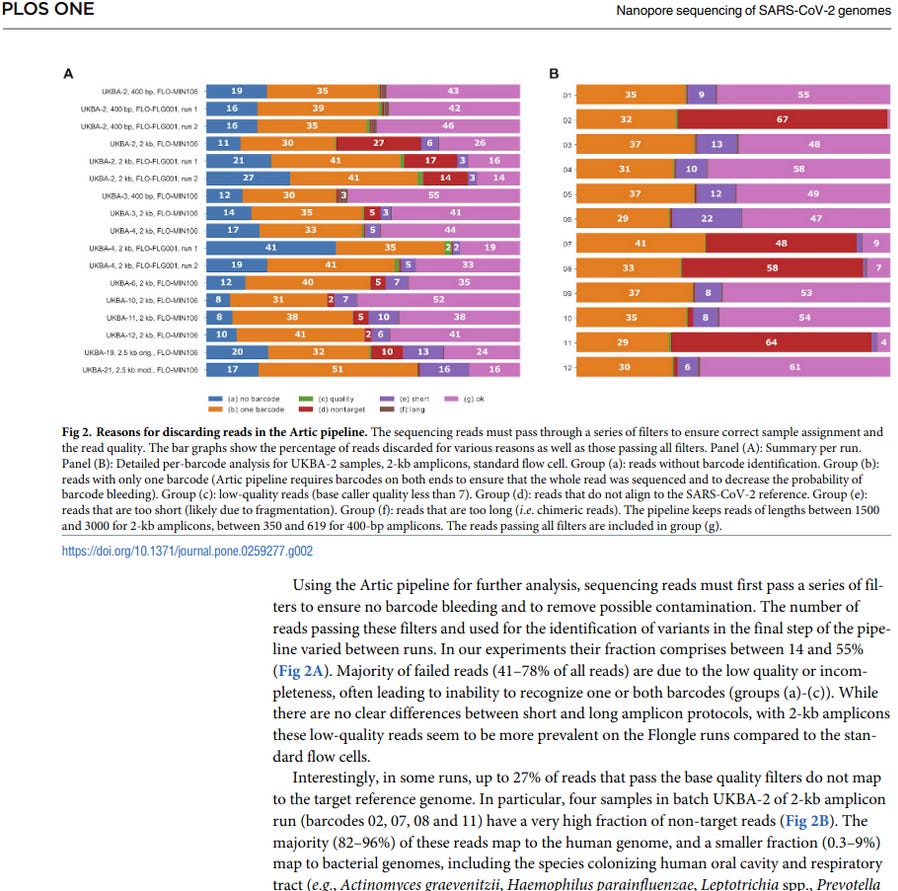
Sequencing of tiled PCR-generated amplicon pools may not be as accurate as one would like. From (Ref. 3):
"Interestingly, in some runs, up to 27% of reads that pass the base quality filters do not map to the target reference genome. In particular, four samples in batch UKBA-2 of 2-kb amplicon run have a very high fraction of non-target reads (Fig 2B). The majority (82–96%) of these reads map to the human genome, and a smaller fraction (0.3–9%) map to bacterial genomes".
"Human and bacterial reads represent artefacts apparently resulting from a non-specific amplification of contaminating nucleic acids present in clinical samples".
Saying “Contaminating nucleic acids present in clinical samples” is a very good way of avoiding mentioning the potential error rate of your measurement based on the selected primers. “Primer design is thus of critical importance for the success of PCR experiments” (Ref. 2). I will keep working on this…
I think it is very important to understand how much variation there is in any measurement, especially if it is going to affect society as a whole in such a dramatic way, as SARS-CoV-2 related measurements have.
ADDITIONAL NOTES ON THE NO-VIRUS PEOPLE:
There are a couple of new videos on this that I think are worth watching, specially the first one with J.C. from https://gigaohmbiological.com. I do not agree with many of the points presented in the second video, however I think it clarifies many confused perspectives on the issue.
Gigaohmbiological & Tim Truth - The Virus Debate - 22 July 22
Is Virology Reasonable? Dr Kevin McCairn Argues SARS-CoV-2 Is Real/ Details Theories On It
My analysis on this can be found in my previous substack [Alternative Hypothesis to Communicable Disease-Causing Agents & Notes on Virology in relation to the No-Virus People]
REFERENCES:
(Ref. 1 - Assembly of SARS-CoV-2 genomes from tiled amplicon Illumina sequencing using Geneious Prime) https://help.geneious.com/hc/en-us/articles/360045070991-Assembly-of-SARS-CoV-2-genomes-from-tiled-amplicon-Illumina-sequencing-using-Geneious-Prime
(Ref. 2 - PCRTiler: automated design of tiled and specific PCR primer pairs) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2896098/
(Ref. 3 - Nanopore sequencing of SARS-CoV-2: Comparison of short and long PCR-tiling amplicon protocols) https://pubmed.ncbi.nlm.nih.gov/34714886/
(VIDEOS)
(GigaohmBiological - Kevin McKernan LIVE: Gigaohm Biological High Resistance Low Noise Information Brief) https://www.twitch.tv/videos/1603141255
(Alternative link to the video - Gigaohm Biological & Kevin
Mckernan Live - 27 Sep 22) https://newtube.app/user/Fredyatelmstreet13/JsYvtSZ
(Gigaohmbiological & Tim Truth - The Virus Debate - 22 July 22) https://newtube.app/user/Fredyatelmstreet13/4nII6BC
(Is Virology Reasonable? Dr Kevin McCairn Argues SARS-CoV-2 Is Real/ Details Theories On It) https://rumble.com/v1mlw6w-is-virology-reasonable-dr-kevin-mccairn-argues-sars-cov-2-is-real-details-t.html